當前位置: 首頁 - 產(chǎn)品專區(qū) - 熱銷產(chǎn)品

H3K27ac polyclonal antibody
| 貨號 | C15410196-10/ C15410196-50 | 售價(元) | 咨詢 |
| 規(guī)格 | 10ug/50ug | CAS號 |
- 產(chǎn)品簡介
- 相關(guān)產(chǎn)品
Polyclonal antibody raised in rabbit against the region of histone H3 containing the acetylated lysine 27 (H3K27ac), using a KLH-conjugated synthetic peptide.
| Lot | A1723-0041D |
|---|---|
| Concentration | 2.8 μg/μl |
| Species reactivity | Human, mouse, rat, Arabidopsis, wide range expected |
| Type | Polyclonal,ChIP grade, ChIP-seq grade |
| Purity | Affinity purified |
| Host | Rabbit |
| Storage Conditions | Store at -20°C; for long storage, store at -80°C. Avoid multiple freeze-thaw cycles. |
| Storage Buffer | PBS containing 0.05% azide and 0.05% ProClin 300. |
| Precautions | This product is for research use only. Not for use in diagnostic or therapeutic procedures. |
| Applications | Suggested dilution | References |
|---|---|---|
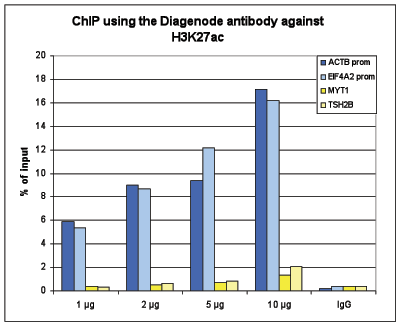
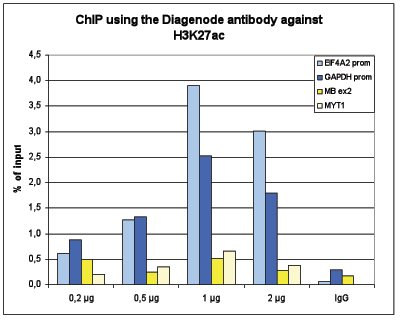
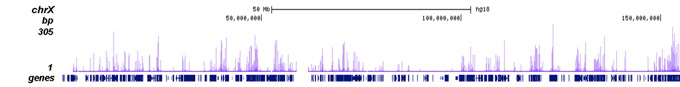
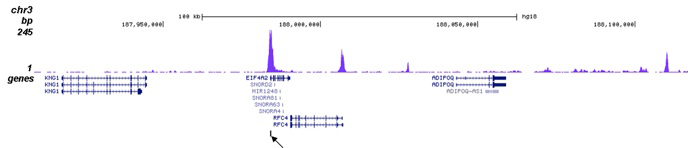
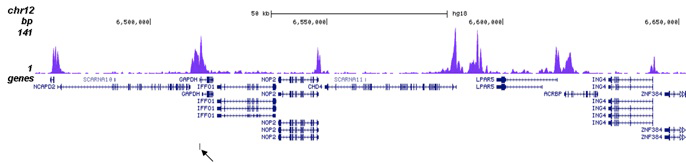
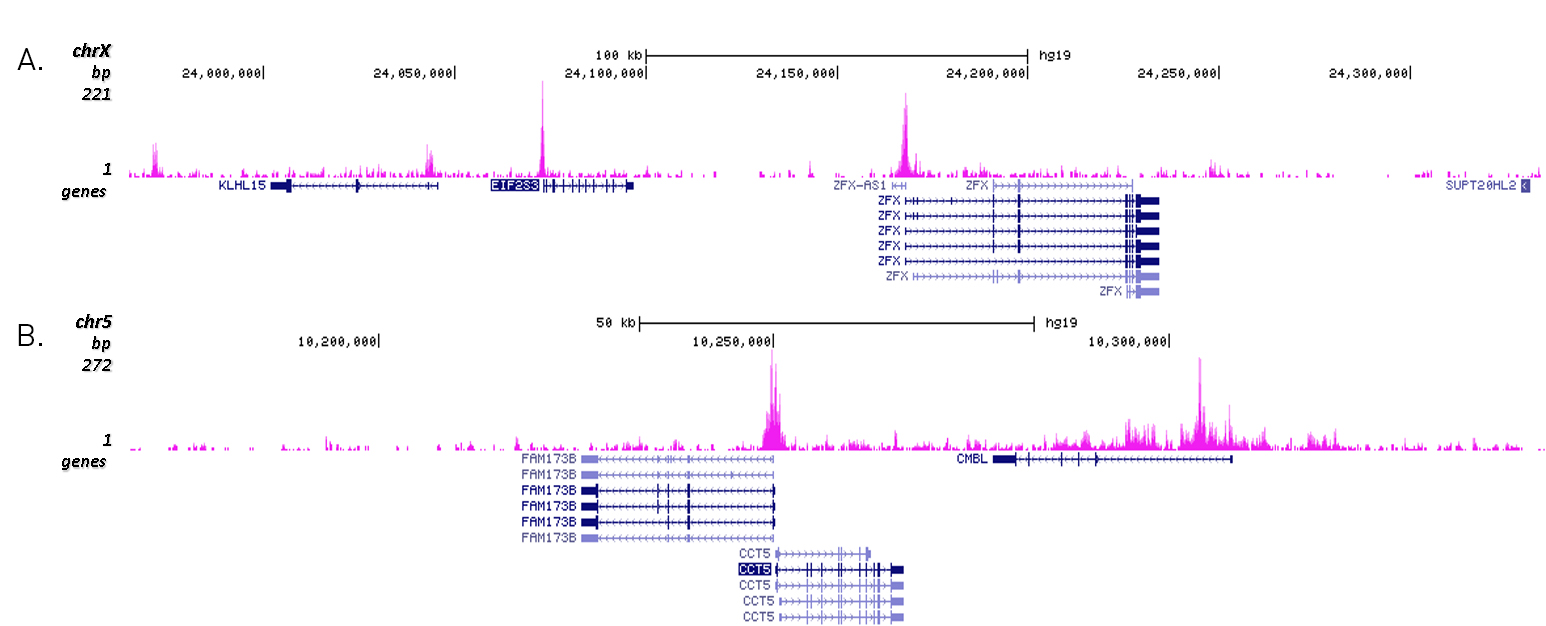
| ChIP/ChIP-seq * | 1 μg/IP | Fig 1, 2 |
| CUT&TAG | 1 μg | Fig 3 |
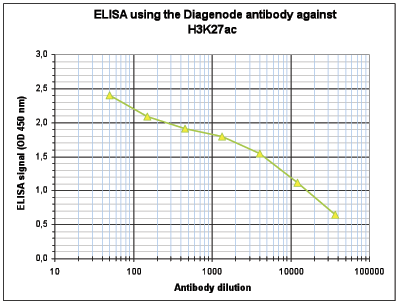
| ELISA | 1:500 | Fig 4 |
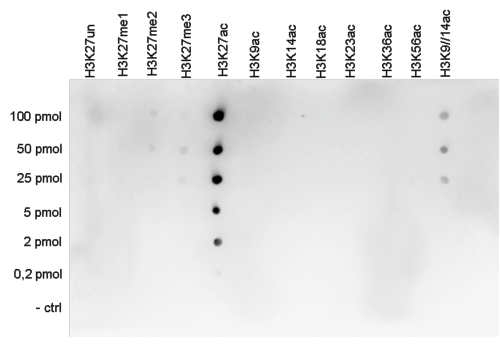
| Dot Blotting | 1:20,000 | Fig 5 |
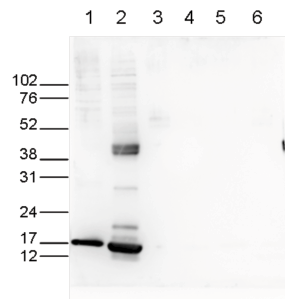
| Western Blotting | 1:1,000 | Fig 6 |
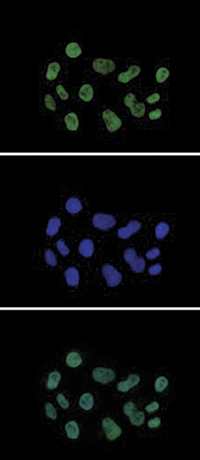
| Immunofluorescence | 1:500 | Fig 7 |
* Please note that the optimal antibody amount per IP should be determined by the end-user. We recommend testing 0.5-5 μg per IP.