H4K20me3 Antibody
| 貨號 |
C15410057-10/C15410057-50 |
售價(元) |
咨詢 |
| 規(guī)格 |
10ug/50ug |
CAS號 |
|
- 產(chǎn)品簡介
- 相關(guān)產(chǎn)品
Polyclonal antibody raised in rabbit against the region of histone H4 containing the trimethylated lysine 20 (H4K20me3), using a KLH-conjugated synthetic peptide.
|
Lot
|
A295-0014P
|
|
Concentration
|
1.07 μg/μl
|
|
Species reactivity
|
Human, mouse
|
|
Type
|
Polyclonal
|
|
Purity
|
Affinity purified
|
|
Host
|
Rabbit
|
|
Precautions
|
This product is for research use only. Not for use in diagnostic or therapeutic procedures.
|
|
Applications
|
Suggested dilution
|
References
|
|
ChIP/ChIP-seq *
|
1-2 μg/ChIP
|
Fig 1, 2
|
|
CUT&TAG
|
1 μg
|
Fig 3
|
|
ELISA
|
1:100
|
Fig 4
|
|
Dot Blotting
|
1:20,000
|
Fig 5
|
|
Western Blotting
|
1:1,000
|
Fig 6
|
|
Immunofluorescence
|
1:300
|
Fig 7
|
* Please note that the optimal antibody amount per IP should be determined by the end-user. We recommend testing 1-5 μg per IP.
-
Validation data
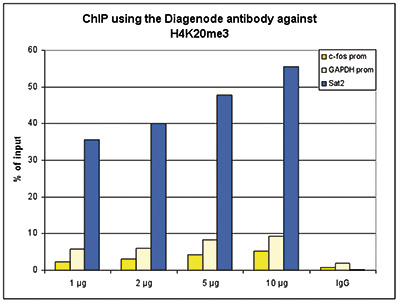
Figure 1. ChIP results obtained with the Diagenode antibody directed against H4K20me3
ChIP assays were performed using human HeLa cells, the Diagenode antibody against H4K20me3 (Cat. No. C15410057) and optimized PCR primer sets for qPCR. ChIP was performed with the “Auto Histone ChIP-seq” kit (Cat. No. C01010022) with sheared chromatin from 1 million cells using the SX-8G IP-Star automated system. A titration of the antibody consisting of 1, 2, 5, and 10 μg per ChIP experiment was analysed. IgG (1 μg/IP) was used as negative IP control. QPCR was performed with primers for promoters of the active genes c-fos (Cat. No. C17011004) and GAPDH (Cat. No. C17011047), used as negative controls, and for the Sat2 satellite repeat region used as a positive control. Figure 1 shows the recovery, expressed as a % of input (the relative amount of immunoprecipitated DNA compared to input DNA after qPCR analysis).
A.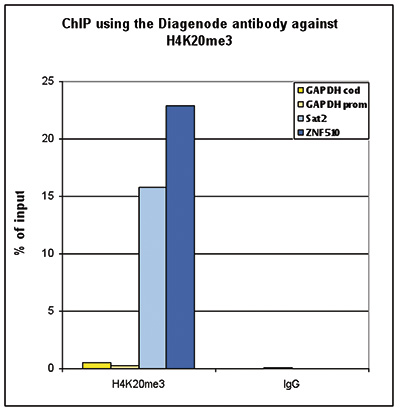
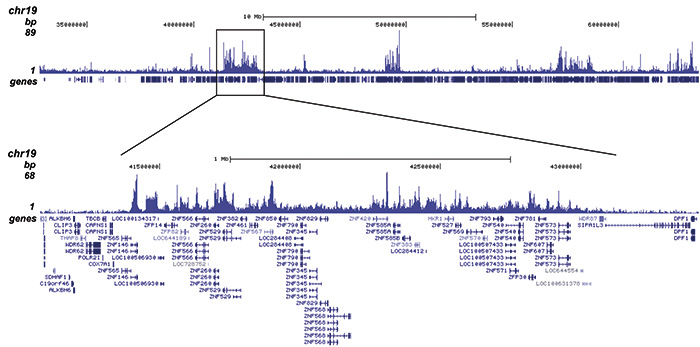
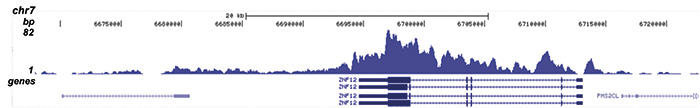
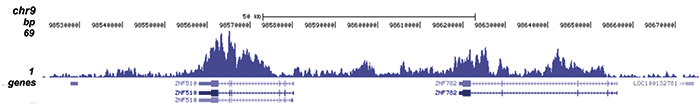
Figure 2. ChIP-seq results obtained with the Diagenode antibody directed against H4K20me3
ChIP was performed with 1 μg of the Diagenode antibody against H4K20me3 (Cat. No. C15410057) on sheared chromatin from 1 million HeLaS3 cells using the “iDeal ChIP-seq” kit. The IP’d DNA was analysed by QPCR with optimized PCR primer pairs for the promoter and coding region of the active GAPDH gene, for the coding region of the ZNF510 gene and for the Sat2 satellite repeat (figure 2A). The IP’d DNA was subsequently analysed on an Illumina Genome Analyzer. Library preparation, cluster generation and sequencing were performed according to the manufacturer’s instructions. The 36 bp tags were aligned to the human genome using the ELAND algorithm. Figure 2B shows the signal distribution along the long arm of chromosome 19 and a zoomin to an enriched region containing several ZNF repeat genes. Figure 2C and D show the enrichment at ZNF12 and ZNF510 on chromosome 7 and 9, respectively. These results clearly show an enrichment of H4K20me3 at ZNF repeat genes.
A.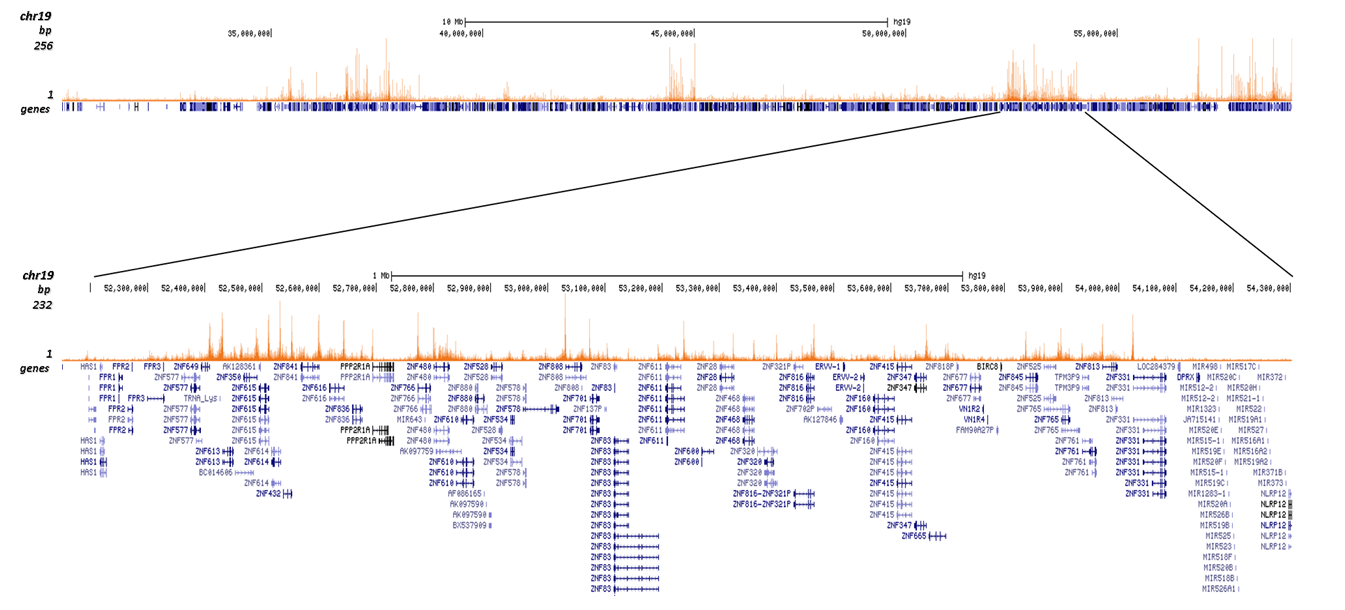
B.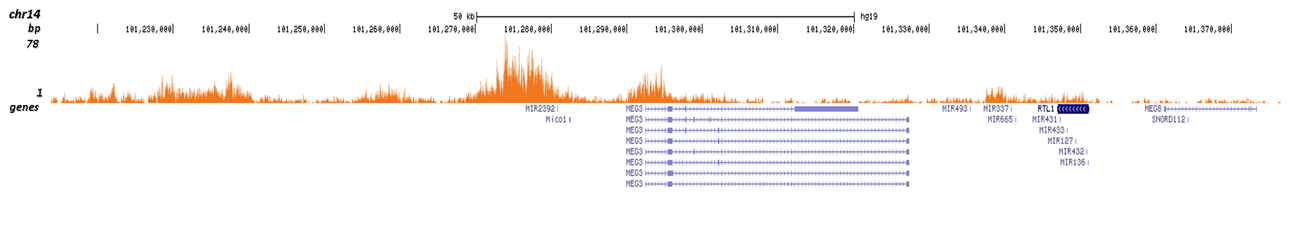
Figure 3. Cut&Tag results obtained with the Diagenode antibody directed against H4K20me3
CUT&TAG (Kaya-Okur, H.S., Nat Commun 10, 1930, 2019) was performed on 50,000 K562 cells using 1 μg of the Diagenode monoclonal antibody against H4K20me3 (cat. No. C15410057) and the Diagenode pA-Tn5 transposase (C01070001). The libraries were subsequently analysed on an Illumina NextSeq 500 sequencer (2x75 paired-end reads) according to the manufacturer's instructions. The tags were aligned to the human genome (hg19) using the BWA algorithm. Figure 3 shows the peak distribution on the long arm of chromosome 19 as well as a zoomin to a region enriched in ZNF repeat genes, and in a genomic region surrounding the MEG3 imprinted control gene on chromosome 14 (figure 3A and B, respectively).
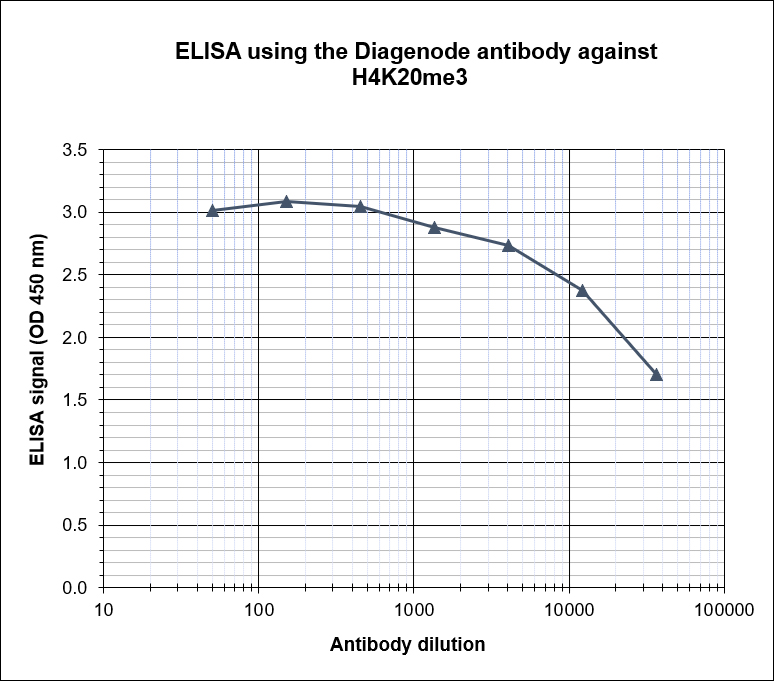
Figure 4. Determination of the antibody titer
To determine the titer of the antibody, an ELISA was performed using a serial dilution of the Diagenode antibody directed against H4K20me3 (Cat. No. C15410057), crude serum and flow through in antigen coated wells. The antigen used was a peptide containing the histone modification of interest. By plotting the absorbance against the antibody dilution (Figure 4), the titer of the antibody was estimated to be 1:7,400.
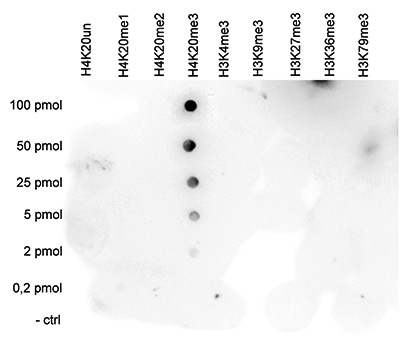
Figure 5. Cross reactivity test using the Diagenode antibody directed against H4K20me3
A Dot Blot analysis was performed to test the cross reactivity of the Diagenode antibody against H4K20me3 (Cat. No. C15410057) with peptides containing other histone modifications and the unmodified H4K20. One hundred to 0.2 pmol of the respective peptides were spotted on a membrane. The antibody was used at a dilution of 1:20,000. Figure 5 shows a high specificity of the antibody for the modification of interest
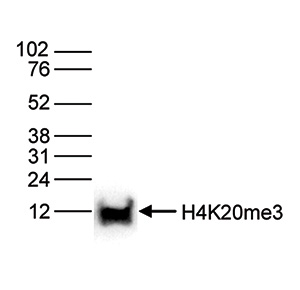
Figure 6. Western blot analysis using the Diagenode antibody directed against H4K20me3
Histone extracts of HeLa cells (15 μg) were analysed by Western blot using the Diagenode antibody against H4K20me3 (Cat. No. C15410057) diluted 1:1,000 in TBS-Tween containing 5% skimmed milk. The position of the protein of interest is indicated on the right; the marker (in kDa) is shown on the left.
A.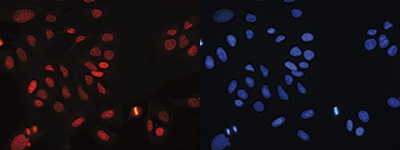
B.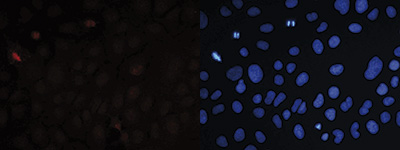
Figure 7. Immunofluorescence using the Diagenode antibody directed against H4K20me3
Human osteosarcoma (U2OS) cells were stained with the Diagenode antibody against H4K20me3 (Cat. No. C15410057) and with DAPI. Cells were fixed with ice cold methanol for 10’ and blocked with PBS/TX-100 containing 5% normal goat serum. Figure 7A: cells were immunofluorescently labeled with the H4K20me3 antibody (left) diluted 1:300 in blocking solution followed by an anti-rabbit antibody conjugated to Alexa568 or with DAPI (right), which specifically labels DNA. Figure 6B: staining of the cells with the H4K20me3 antibody after incubation of the antibody with blocking peptide (Cat. No. C16000057), concentration: 5 ng/μl).