
EZ-Tn5? Insertion Kit/EZ-Tn5? 插入試劑盒
| 貨號 | EZI982K | 售價(元) | 6950 |
| 規(guī)格 | 10 rxns | CAS號 |
- 產品簡介
- 相關產品
產品信息
|
貨號 |
產品名稱 |
規(guī)格 |
價格/元 |
|
TNP92110 |
EZ-Tn5 Transposase/EZ-Tn5轉座酶 |
10 units |
6418 |
|
EZI982K |
EZ-Tn5? <KAN-2> Insertion Kit/EZ-Tn5? <KAN-2>插入試劑盒 |
10 rxns |
6950 |
|
EZI921T |
EZ-Tn5? <TET-1> Insertion Kit EZ-Tn5? <TET-1>插入試劑盒 |
10 rxns |
6999 |
|
EZI912D |
EZ-Tn5? <DHFR-1> Insertion Kit/EZ-Tn5? <DHFR-1>插入試劑盒 |
10 rxns |
7060 |
|
EZI03T7 |
EZ-Tn5? <T7/KAN-2> Promoter Insertion Kit/EZ-Tn5? <T7/KAN-2>啟動子插入試劑盒 |
10 rxns |
7312 |
|
EZI011RK |
EZ-Tn5? <R6Kγori/KAN-2> Insertion Kit/EZ-Tn5? <R6Kγori/KAN-2>插入試劑盒 |
10 rxns |
7517 |
|
CIS40025 |
CopyCutter? Induction Solution/CopyCutter?誘導試劑 |
25 mL |
856 |
Transposons are mobile DNA sequences found in the genomes of prokaryotes and eukaryotes. Transposon tagging has long been recognized as a powerful research tool for randomly distributing primer binding sites, creating gene“knockouts”, and introducing a physical tag or a genetic tag into large target DNAs. One frequently used transposition system is the Tn5 system isolated from gram-negative bacteria. Though a naturally occurring transposition system, the Tn5 system can be readily adapted for routine use in research laboratories for the following reasons:
1) Tn5 transposase is a small, single subunit enzyme that has been cloned and purified to high specific activity.
2) Tn5 transposase carries out transposition without the need for host cell factors.
3) Tn5 transposon insertions into target DNA are highly random.
4) Tn5 transposition proceeds by a simple“cut and paste”process. Although the chemistry is unique, the result is similar to using a restriction endonuclease, with random sequence specifi city, accompanied by a DNA ligase activity.
5) Tn5 transposase will transpose any DNA sequence contained between its short 9 basepair Mo saic End (ME) Tn5 transposase recognition sequences.
In 1998 Goryshin and Reznikoff1demonstrated that a fully functional Tn5 transposition system could be reconstituted in vitro. Additionally, the transposition efficiency of this system has been increased more than 1,000-fold compared to wild-type Tn5 by introducing mutations in the transposase gene and in the 19-bp Tn5 ME transposase recognition sequence.
Lucigen’s EZ-Tn5 Transposon Tools (kits and reagents) are based on the hyperactive Tn5 transposition system developed by Goryshin and Reznikoff.
優(yōu)點:
Insert a kanamycin, tetracycline, or DHFR selectable marker into any DNA sequence in vitro
Skip primer walking - simplify Sanger sequencing of large DNA inserts
Speed functional analysis without subcloning - create libraries of random mutants from purified DNA
Minimize insertion bias with the hyperactive Tn5 system, known for highest level of randomness
組成成分:
EZ-Tn5? Transposase 10 U :儲存在-20℃
EZ-Tn5? <R6Kγori/KAN-2> Transposon:儲存在-20℃
EZ-Tn5? 10X Reaction Buffer:儲存在-20℃
EZ-Tn5? 10X Stop Solution:儲存在-20℃
KAN-2 FP-1 Forward Primer:儲存在-20℃
R6KAN-2 RP-1 Reverse Primer:儲存在-20℃
pUC19/3.4 Control Target DNA:儲存在-20℃
Sterile Water:儲存在-20℃
數據:
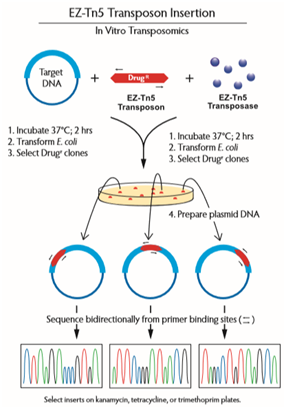
Figure 1. The process for generating DNA sequencing templates using an EZ-Tn5 Insertion kit.
技術參數
產品優(yōu)點- Insert a kanamycin, tetracycline, or DHFR selectable marker into any DNA sequence in vitro
- Skip primer walking - simplify Sanger sequencing of large DNA inserts
- Speed functional analysis without subcloning - create libraries of random mutants from purified DNA
- Minimize insertion bias with the hyperactive Tn5 system, known for highest level of randomness
產品應用- Faster sequencing of large DNA molecules, as compared to primer walking, random subcloning, or generating nested deletions with exonuclease III and mung bean nuclease.
- Making insertion mutants or gene “knockouts” in vitro.
- Introducing a kanamycin resistance selection marker into any DNA.


